I'm taking a break from my Summer break
to post a few interesting papers that have come out within the past
couple of months.
This paper supports such a notion of
continuous gene-flow between Africans and non-Africans since the
major Out of Africa event that was precursor to the populating of all
continents outside of Africa.
To be sure, such a notion is not new
but has been highlighted before by methods used by authors such as Li and Durbin (2011) for instance. Such a notion, is also sufficient to
explain the intermediate genetic nature of West Eurasians, I.e
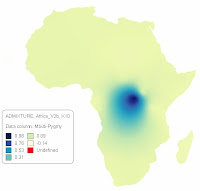
between Africans and East Asian/Native Americans, that I have blogged about and demonstrated using ADMIXTURE in the past.
A few quotes from the paper:
"In
this paper, we study the length distribution of tracts of identity by
state (IBS), which are the gaps between pairwise differences in an
alignment of two DNA sequences. These tract lengths contain
information about the amount of genetic diversity that existed at
various times in the history of a species and can therefore be used
to estimate past population sizes. IBS tracts shared between DNA
sequences from different populations also contain information about
population divergence and past gene flow. By
looking at IBS tracts shared within Africans and Europeans, as well
as between the two groups, we infer that the two groups diverged in a
complex way over more than 40,000 years, exchanging DNA as recently
as 12,000 years ago."
"To
illustrate the power of our method, we use it to infer a joint
history of Europeans and Africans from the high coverage 1000 Genomes
trio parents. Previous analyses agree that Europeans experienced an
out-of-Africa bottleneck and recent population growth, but other
aspects of the divergence are contested [47].
In one analysis, Li and Durbin separately estimate population
histories of Europeans, Asians, and Africans and observe that the
African and non-African histories begin to look different from each
other about 100,000–120,000 years ago; at the same time, they argue
that substantial migration between Africa and Eurasia occurred as
recently as 20,000 years ago and that the out-of-Africa bottleneck
occurred near the end of the migration period, about 20,000–40,000
years ago. In contrast, Gronau, et
al. use
a likelihood analysis of many short loci to infer a Eurasian-African
split that is recent enough (50 kya) to coincide with the start of
the out of Africa bottleneck, detecting no evidence of recent gene
flow between Africans and non-Africans [14].
The older Schaffner, et
al. demographic
model contains no recent European-African gene flow either [48],
but Gutenkunst,et
al. and
Gravel, et
al. use
SFS data to infer divergence times and gene flow levels that are
intermediate between these two extremes [22], [49].
We aim to contribute to this discourse by using IBS tract lengths to
study the same class of complex demographic models employed by
Gutenkunst, et
al. and
Gronau, et
al.,
models that have only been previously used to study allele
frequencies and short haplotypes that are assumed not to recombine.
Our method is the first to use these models in conjunction with
haplotype-sharing information similar to what is used by the PSMC and
other coalescent HMMs, fitting complex, high-resolution demographic
models to an equally high-resolution summary of genetic data."
"We
estimate that the European-African divergence occurred 55 kya and
that gene flow continued until 13 kya. About 5.8% of European genetic
material is derived from a ghost population that diverged 420 kya
from the ancestors of modern humans. The out-of-Africa bottleneck
period, where the European effective population size is only 1,530,
lasts until 5.9 kya."
"Our
inferred human history mirrors several controversial features of the
history inferred by Li and Durbin from whole genome sequence data: a
post-divergence African population size reduction, a sustained period
of gene flow between Europeans and Yorubans, and a “bump” period
when the ancestral human population size increased and then decreased
again. Unlike Li and Durbin, we do not infer that either population
increased in size between 30 and 100 kya. Li and Durbin postulate
that this size increase might reflect admixture between the two
populations rather than a true increase in effective population size;
since our method is able to model this gene flow directly, it makes
sense that no size increase is necessary to fit the data. In
contrast, it is possible that the size increase we infer between 240
kya and 480 kya is a signature of gene flow among ancestral hominids."
"Our
estimated divergence time of 55 kya is very close to estimates
published by Gravel, et
al.and
Gronau, et
al.,
who use very different methods but similar estimated mutation rates
to the  per
site per generation that we use in this paper. However, recent
studies of de
novo mutation
in trios have shown that the mutation rate may be closer to
per
site per generation that we use in this paper. However, recent
studies of de
novo mutation
in trios have shown that the mutation rate may be closer to  per
site per generation [51], [55], [56].
We would estimate older divergence and gene flow times
(perhaps
per
site per generation [51], [55], [56].
We would estimate older divergence and gene flow times
(perhaps  times
older) if we used the lower, more recently estimated mutation rate.
This is because the lengths of the longest IBS tracts shared between
populations should be approximately exponentially distributed with
decay rate
times
older) if we used the lower, more recently estimated mutation rate.
This is because the lengths of the longest IBS tracts shared between
populations should be approximately exponentially distributed with
decay rate  ."
."
(2)
Genetic
and archaeological perspectives on the initial modern human
colonization of southern Asia (Closed Access)
This paper
discusses some points, rather the lack of evidence, that makes a
pre-toba migration of modern humans outside of Africa almost impossible to reconcile with currently
available evidence.
A few quotes from the paper:
"There
are currently two sharply conflicting models for the earliest modern
human colonization of South Asia, with radically different
implications for the interpretation of the associated genetic and
archaeological evidence (Fig. 1). The first is that modern humans
arrived ∼50–60 ka, as part of a generalized Eurasian dispersal of
anatomically modern humans, which spread (initially as a very small
group) from a region of eastern Africa across the mouth of the Red
Sea and expanded rapidly around the coastlines of southern and
Southeast Asia, to reach Australia by ∼45–50 ka (7–10, 14–18)
(Fig. 2). The second, more recently proposed view, is that there was
a much earlier dispersal of modern humans from Africa sometime before
74 ka (and conceivably as early as 120–130ka), reaching southern
Asia before the time of the volcanic “supereruption” of Mount
Toba in Sumatra (the largest volcanic eruption of the past 2 million
y) at ∼74 ka (1–6)."
"We
find no evidence, either genetic or archaeological, for a very early
modern human colonization of South Asia, before the Toba eruption.
All of the available evidence supports a much later colonization beginning
∼50–55 ka, carrying mitochondrial L3 and Y chromosome C, D, and F
lineages from eastern Africa, along with the Howiesons Poort-like
microlithic technologies (see above and Genetics and Archaeology). We
see no reason to believe that the initial modern human colonization
of South and Southeast Asia was distinct from the process that is now
well documented for effectively all of the other regions of Eurasia
from ∼60 ka onward, even if the technological associations of these
expanding populations differed (most probably for environmental
reasons) between the eastern and northwestern ranges of the
geographical dispersal routes."
"The
archaeological evidence initially advanced to support an earlier
(pre-Toba) dispersal of African-derived populations to southern Asia
has since been withdrawn by the author responsible for the original
lithic analyses, who now suggests that they are most likely “the
work of an unidentified population of archaic people” (ref. 11, p.
26). Meanwhile, the genetic evidence outlined earlier indicates that
any populations dispersing from Africa before 74 ka would predate the
emergence of the mtDNA L3 haplogroup, the source for all known,
extant maternal lineages in Eurasia (8, 28) (Fig. 5). The size of the
mtDNA database is very substantial: currently there are almost 13,000
complete non-African mtDNA genomes available, not one of which is
pre-L3."
This
paper, written by a geneaolgoical community member, has made an impressive effort at
creating and automating a comprehensive method to pylogenetically classify Geno 2.0
YDNA SNPs. Details of the algorithm are not available:
"To
illustrate this, the author has used this Y-tree clade predictor
(using the latest ISOGG tree as a basis for comparison) to classify
over 1650 sets of publicly accessible Geno 2.0 Y-SNP calls. This
information was then used as an input into another algorithm designed
by the author – an algorithm developed to automate the construction
of a phylogenetic Y-tree, while overcoming the challenges identified
above. The technical details of this process will remain proprietary
for the time being."