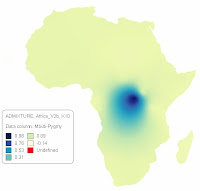
Below is a contour map representing the African ADMIXTURE component at K=2 for the Global data set (V2) which can be downloaded here, and population specific percentages that can be seen here.
Contour map generated using Mapviewer7, Kriging method was used for gridding. ADMIXTURE outputs for all New World, Jewish, Singapore-Chinese and Singapore-Indian populations were removed before the generation of the map.
 |
| African cline from ADMIXTURE, K=2 . Black dots represent locations of sampled populations |
Some things to note,
- Since this is a K2 run, the OOA or the 'other' component has a complete mirror distribution relative to the distribution of the African component seen in the above.
- The regions where the brown color dominates (20-35% African ) are the same regions that are later on absorbed by the new component that arises @ K=3, which finds its peaks in West Eurasians and has an FST that is intermediate between those of the African and East Asian/Amerindian components.
- It is notable to observe the congruence of the above with the distribution of global genetic as well as phenotypic diversity (below)1
 |
| Global phenotypic and genetic Diversity |