I will attempt to highlight the main changes.
First, in the begining of last year Trombetta et. al came out with the paper:
A New Topology of the Human Y Chromosome Haplogroup E1b1 (E-P2) Revealed through the Use of Newly Characterized Binary Polymorphisms
This paper not only found new SNPs and structure below E1b1b1 (E-M35), but also revamped the overall E1b1 (E-P2) structure as seen below :
More over, the paper further confirmed what was previously thought about the origin of E1b1 (E-P2) in East Africa by stating:
"The new topology here reported has important implications as to the origins of the haplogroup E1b1. Using the principle of the phylogeographic parsimony, the resolution of the E1b1b trifurcation in favor of a common ancestor of E-M2 and E-M329 strongly supports the hypothesis that haplogroup E1b1 originated in eastern Africa, as previously suggested, and that chromosomes E-M2, so frequently observed in sub-Saharan Africa, trace their descent to a common ancestor present in eastern Africa."
The paper then went further to test the New SNPs on the old Cruciani '04 dataset , see below for the East African results once the New SNPs were incorporated:
Note that E-V42 and E-V92 were not found outside of Ethiopia, and that newly restructured 'sibling' of E-M35 , i.e. E-M281, found in the 34 Ethiopian Amhara sample (previously recognized as E-M215*).
Accordingly, SNPs from Haplogroup E tested in Ethiopia to date by the various papers published has been updated below. Note that Trombetta '11 is not itemized separately since the dataset is the same as that from Cruciani '04.
The next update for the E-M35 phylogeny came at the end of last year, however this time not in a form of a published paper but by the work of individuals in the genealogical community, by data mining Y-chromosomes from the 1000 genome project a couple of SNPs that changed the internal structure of E-M35 were found. The first SNP, labeled as Z827, united E-M123, E-V257, E-V42 and E-M293. The second SNP, labeled as Z830 found to be downstream of E-Z827, united E-M123, E-M293 and E-V42. The newly proposed structure is shown below, it is important to note that more samples need to be tested to verify this new structure.
This below is therefore an attempt at the phylogeographic route of E-M35 and its various major and minor subclades since its inception.



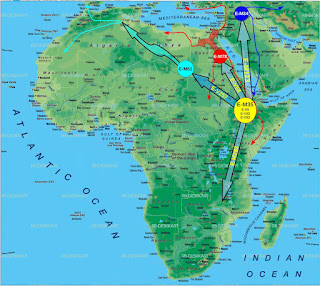



What role, if any, would you give to Sudan?, which is obligate corridor for most of this flows and, at least in my mind, alternative or shared possible origin for E1b1b to Ethiopia. Also, related, within Ethiopia, how much of this diversity is in the tribal province of the SW (if you know at all)?
ReplyDeleteAnyhow, and following the data of Semino 2004 I'd say that E1b1b1b1-M81, looks totally like coalescing in NW Africa, maybe in Morocco and not in the Lybian Desert, where you placed it a bit happily. Whether that happened in relation with this or that Paleolithic culture and related migration is of course arguable but there is almost no E-M81 outside NW Africa, regardless that E1b1b1b-V257 probably coalesced in East Africa (Nile area), like the rest of the family.
A totally different case is that of E1b1b1a1-M78, which looks (again in Semino'04) split between the Horn and NW Africa, with important ramification towards Egypt and even SE Europe, this looks to me more like E1b1b1-M35 as a whole, i.e. original from Ethiopia or Sudan and then scattered around the Mediterranean mostly. So these two are either the same wave or, more likely IMO, two separate waves but from the same area.
Do you have any data that could challenge this perception of mine?
Hi Maju, first let me say I haven't forgotten about the matlab code I said I'd email you, I just haven't logged on to my UNIX station yet.
ReplyDeleteNow, with respect to the map, it is a very general map based on information I have gathered on E1b1b thus far, by no means is it set in stone, it is the most reasonable migration route based on spatial frequencies of the E1b1b family that we have.
With respect to Sudan vis-a-vis Ethiopia, thus far we have no major E1b1b subclades from that region except E-M78. Where as Ethiopia thus far has the subclades of E-M35 that are only found there: E-V42, E-V92, E-V6 and off course one that is a sister to E-M35 and downstream of E-M215 and that is E-M281. This is what makes Ethiopia a better candidate than Sudan for the origin of E-M35, and likely also the origin of E-M35's ancestor E-P2(E1b1), that is by far the dominating lineage (>70%) of the Entire African continent.
With regards to the coalescing of E-M81, you may be correct, but I point you to the data from the El-Hayez Oasis in the Egyptian western desert, almost half of the E lineages found there belonged to E-M81, I forgot the papers name I just have the graphic I made of it years ago, I'll dig deeper and find you the paper.
With respect to within Ethiopia distribution of E1b1b1, be it SW/SE/NW or NE, the only detailed data we have is the cruciani data set that you can find the results for in the post of this thead, I used the center of Ethiopia because different subclades of E1b1b and ancestral E1b1b1* is found pretty much in the entire country amongst populations living both North and South (Wolayta, Amhara, Oromo, Beta Israel et. al)
Lastly, E-M78 looks like it coalesced in Egypt/Sudan or maybe even as far west close to the Libyan border, most of the major subclades of E-M78 are also found there, most importantly E-V12*, which later went back south largely in the form of E-V32, also E-V65, E-V13 are not found in the Ethiopian area, there is a little bit of E-V22 however.
Ok found the paper on E-M81 in Egypt I mentioned earlier, I just have the link to the Abstract and the frequencies however: http://www.ncbi.nlm.nih.gov/pubmed/19425100
ReplyDeleteThey found the following:
J1-M267 31%
E-M78 29%
E-M81 29%
E-M2(x M58) 6%
E-M35(x M78,M81,M123) 6%
The oasis is close to Al-Bawiti in the Northwest of the country.
Sorry I did not come back before: I forgot to subscribe it seems.
ReplyDeleteRe. the Western Desert Oasis, it's hard to accept as evidence, as it could well have been deserted and repopulated a thousand times for what we can know. I'd rather consider the lineages found in more densely populated areas like coastal Libya or even Fezzan (or Egypt itself but the banks of the Nile). It's just too easy for a group of Berbers to have backmigrated in many different occasions, for example in the periods when Ancient Egypt felt threatened by the Libu or whatever.
As for Sudan, I take your word but I must say that I am very perplex because, after all, Ethiopia and all the Horn only communicate with the rest of Africa by the Sudans, Kenya and maybe the sea, what makes former Sudan an obligate transit and a place where I would expect to find at least some high level of diversity even if it'd be second to Ethiopia or whatever.
I published back in the day this map made by Argiedude (who is a very dedicated aficionado to population genetics) of African Y-DNA. By now the sources are all lost (though guess they can be requested if need be: I still get emails from Argiedude now and then and he may keep the database) and the nomenclature has become a bit obsolete, but I still discern something more than just E-M78 (more precisely E-V32 in South Sudan): E-M293 in South Sudan and lots of DE* (unclassified YAP+) in all three Sudanese regions. True that it is not very informative but most of the information you detail here was back then (2.5 years ago!) also amorphous DE*.
So I guess that at leas a fraction of the diversity found now in Ethiopia is also present in the Sudans. Unlike Sudan (and back then also Ethiopia), other African areas have only very few unclassified YAP+ (main exception being Westernmost Africa: Senegal, Gambia and Guinea-Bissau).
But guess we will have to wait until the diversity hidden under the YAP+ marker in the Sudans is clarified.
"and ancestral E1b1b1*"...
Unclassified is not the same as "ancestral". You can make the "asterisk" paraphyletic clade to weight as one or several well defined lineages (depending on what you think that most likely hides behind it) but is not ancestral. In mtDNA sometimes we may find underived lines but that does just not happen in Y-DNA: just that researchers have not yet found the defining mutations under that node (but they are there for sure).
I guess it's not important for this case (because you also mention "different subclades of E1b1b") but it is an important general notion to keep in mind.
Anyhow, a pleasure to debate this.
Hi Maju, I am familiar with that YDNA map made by argeidude, while a decent piece of work, however the labeling of the yellow portion of the pie chart can be misleading if you do not dig into the data yourself. I have already checked the Ethiopian data,and corrected it, I am not sure how the other DE slices need to be resolved without looking at all of them in detail, but here is how the Ethiopian data needs to be resolved (not updated for post 2009 SNPs or papers):
ReplyDelete1) Haplogroup J should be at an average frequency of ~ 18.3 % (instead of 20%)
2) Haplogroup B should be at an average frequency of ~ 2.8% (instead of 1%)
3) Haplogroup E2 should be at an average frequency of ~3.7% (instead of 2%)
4) Haplogroup E1b1a should be at an average frequency of ~0.65% (instead of 0%)
5) Haplogroup E* should be added at an average frequency of ~0.65%
6) The breakdown of the DE(x E1a, E1b1a,V32,M293,E2) portion of the Ethiopian pie should be detailed out as follows:
E1b1b1c / E-M123 ~ 5.2%
E1b1b1a / E-M78 (x V32) ~ 7% (Probably most, if not all E-V22)
E1b1b1d / E-M281 ~0.4%
E1b1b/E1b1b1* = E-M215/M35* (x E-M293, E-M123, E-M81,E-M78, E-M281) ~ 9% (Some could be E-V6 or P-72)
E1b1* = E-Pn2* (x E-M35, E-M2) ~ 12%
Thus, the least common denominating phylogenetic node should be labeled as E1b1(x E1a, E1b1a,V32,M293,E2) for Ethiopia and not DE(x....), since no Haplogroup D or DE* has ever been found in Ethiopia while all the remaining haplotypes in the yellow cluster belong downstream of E1b1 (E-PN2).
I see. While most of the other figures are similar to those of Argiedude (small variation, proper of any two distinct samples or sets of samples), those under DE* have been discerned. That's why I wonder if at least some of this diversity is also found in Sudan, also under Argie's DE* label.
ReplyDeleteAnyhow, while no DE* has yet been found in Ethiopia (or anywhere outside of West Africa and Tibet), that does not discard the possibility of being some, unless all is also positively tested for E (or E-derived) markers. Not sure if that has been the case...
Yes maju, I have a running compilation of all YDNA studies done on Ethiopia to date,and it is a clear error to label that yellow slice as DE(X...), since all the haplotypes are below E1b1 (E-P2), 2/3 of them have been tested to belong to some type of E1b1b1 and the remaining 1/3 have been tested to belong to E1b1(X E-M2, E-M35).
ReplyDeleteAs far as the Sudanese Data, almost all of it comes from Hassan et. al 2008, all the DE(X...) in that slice belongs to E1b1b (X M78), which leaves you with a wide variety of E1b1b subtypes those haplotypes could belong to like: E-M81, E-V68*, E-M123, etc....
... "and it is a clear error to label that yellow slice as DE(X...), since all the haplotypes are below E1b1 (E-P2), 2/3 of them have been tested to belong to some type of E1b1b1 and the remaining 1/3 have been tested to belong to E1b1(X E-M2, E-M35)".
ReplyDeleteThen fair enough.
"As far as the Sudanese Data, almost all of it comes from Hassan et. al 2008, all the DE(X...) in that slice belongs to E1b1b (X M78)"...
Alright. Thanks for the info.
Guess I'm always hoping that some DE(xD,E) will be found some day in East Africa but guess not. Same as CDEF*... nowhere to be found anymore. Enfin...
You are welcome. One more piece of data that may further support my speculation on E-M81's coalescence being closer to the Egypt/Libyan/Chad/Sudan border rather than Coastal Northwest Africa may also be taken from Underhill et. al 2000, this paper found 5% E-M81(X M107,M165) in 40 individuals sampled from Sudan. Off course, like you said this area is one of the least densely populated areas in Africa, it is basically uninhabited today, but has it always been like this?
ReplyDelete"but has it always been like this?"
ReplyDeleteObviously not "always", because there have been pluvial episodes but otherwise the oases are not a good reference, really.
I think that there is a potential to discern the fine detail of internal diversity within the clade and such. I do not have the data for E-M81 but we do have some good data for mtDNA U6.
Interestingly, the main compiler of this data, Nicole Maca-Meyer, proposed a Sudanese (or otherwise "Nubian") origin for the haplogroup. However looking at the raw data that claim can't be sustained because most of the basal diversity of U6 is in Morocco (followed by Canary Islands and Iberian Peninsula), so even if she is correct in U6a having experienced a (secondary) flow from East to West together with some U6*, this cannot be the origin.
On light of that and also oldest Oranian dates for the Moroccan sites, I conclude that there must have been at least one expansion from NW Africa eastward (in relation with Oranian surely), even if there was later a backflow from Sudan/Nubia (Capsian culture and related lineages).
This can well confuse the matter quite a bit (and never mind the Megalithic cultural flow surely related to the 'Libu', Berber, pressure on Egypt in the Bronze Age, which can imply a second genetic flow from the West, at least hypothetically).
Whatever the case, in all this picture the low aridity period is limited and maybe not too relevant (the dates don't seem to match any known cultural flow).
E-M78 originated in the Horn of Africa, not North Africa. In autosomal genetic studies Egyptians show a lot of Horn African genetic input while the reverse (Egypto-Nubian autosomal input) in the Horn is nonexistent. Perhaps E-M78 used to be more diverse in the past in the Horn but many sub-clades could have died out due to ecological upheavals.
ReplyDelete"Perhaps E-M78 used to be more diverse in the past in the Horn but many sub-clades could have died out due to ecological upheavals."
ReplyDeletePerhaps, but that doesn't explain why a large number of E-V12*(X V32, M224) ancestral to E-V32 (which is what is mostly found in the horn), is only found in southern Egypt. Current data points to E-M78 originating in Southern Egypt / Northern Sudan, Aswan or Lake Nasser vicinity, it is hard to speculate on what could have happened when you have very little empirical data to back up such a speculative scenario with.
http://4.bp.blogspot.com/_uqlFudEVY8U/S-Gz54CtgCI/AAAAAAAAAEc/hu1eqSyp9hk/s1600/E-M78_North_East_Africa2.PNG
The main problem with finding more diverse E-M78 samples in the Horn is that researchers sample the same groups all the time. For example, the main Y-DNA studies on Somalis sampled diaspora people living in Western countries who are almost all Hawiye and Darod (patriarchal tribes lacking Y-DNA diversity). I am sure if they focused more on minor/outcast Somali tribes like the Midgaan, Gabooye, Eeyle, Digil, Mirifle etc they would find a lot more Y-DNA diversity and perhaps place the origin of E-M78 in the Horn. I mean who would have thought that the Borana carried the basal E1b1b1b* (E-V257) clade?
ReplyDeleteWhat is so surprising about finding ancestral E-V257* in the borena when the lineage's most immediate ancestors (E-P2-->E-M215-->E-M35-->E-Z827-->E-Z830) were all likely born very close by?
ReplyDeleteAs far as your speculation that more E-M78 subclades will be found in Somalis when a rather more diverse set of Somalis are sampled than currently are, I simply can not subscribe to, Ancestral E-V68* may be found but likely from what we know today not many subclades of E-M78 other than E-V32 and E-V22 (to a significantly smaller extent) will be found.
Well, Z830 hasn't been observed in the region so far IIRC, so therefore it's surprising.
ReplyDeleteThe reason why I am skeptical of the putative Northern/Saharan origin of E-M78 is that it does not align with autosomal results obtained from the Horn region. Most of the populations in Northern/Saharan Africa are characterized by West Asian and Central African autosomal input, as can be seen here:
http://3.bp.blogspot.com/-m5emkUFdgys/TgRIBluaAmI/AAAAAAAAAic/i3y0AGPK3Gw/s1600/ADMIXTURE_10.png
This is not observed in the Horn, therefore casting doubt on the Northern/Saharan origin. However, there is also the possibility that modern Saharan populations are not very representative of their early neolithic ancestors.
Neither Z827 or Z830 have been observed in the region so far, these are very recent SNPs that do not even appear in published papers, but from the down stream SNPs that they unite we can make some reasonable inferences.
ReplyDeleteAutosomal data is ambiguous and does not describe phylogeny:
"Most importantly, it should be emphasized that this approach does not model phylogeny. Therefore caution needs to be exercised in drawing inferences regarding the meaning of genetic difference and similarity. For example, if two or more different components (putative ancestry signals) are present in a sample, it is not possible to differentiate the relative contribution of recent admixture versus shared ancestry to population structure."
.......
"Therefore, the term ‘ancestry’ as inferred from structure -like analyses used in the current study, refers to genetic relatedness and should be considered in terms of genetic ‘similarity’ and ‘dissimilarity’ irrespective of its genesis, and does not reflect phylogenetic history, though might mirror it."
Behar et. al (2010)
http://discovery.ucl.ac.uk/1310136/1/1310136.pdf
ReplyDeleteData on the Nilo-Saharan Anuak in Ethiopia...
Dont know how long they been in Ethiopia.
I dont know if you want to include this or not but I know you collect data so.
-Beyoku.
Thanks for the paper, looks extensive, I have always wanted a Nilo-Saharan data-set to add as a separate bar stack to my collation of Ethiopian YDNA chart , (as well as omotic speakers), but the resolution of this paper doesn't seem to fit the YDNA tree resolution I have in my chart.
DeleteIt seems like 108 Anuaks were tested for BT*(x DE,KT) ~25%, E*(xE1b1a1) ~17%, A3b2~19%, E1b1a1a1f1a (M191/P86) ~39%.
Since the 'E1b1a' the paper is reporting would be actually E1b1a1 aka M2/SY81 in the newer nomenclature, this means that the E*(xE1b1a1) can me anything, including E-V38*(E1b1a*), or E-M329 (E1b1a2), some type of E1b1b or even E* . The BT*(xDE,KT) , can be a lot of things that is neither K-M9 nor YAP+, but I'm guessing most would be B? This would have been a great opportunity to add Nhilo Saharan speakers from Ethiopia as a separate column in my collation but the resolution is just too low. Thanks for the heads up anyway.
What does the (x) indicate in E1b1a1
DeleteThe "x" generally means "eXcepted" whatever follows, for example "E-Pn2* (x E-M35, E-M2)", means that M35 and M2 subclades are eXcluded from the E-Pn2 category in that specific text.
DeleteThe "*" means "others", with meaning sometimes implicit or, in the example, explicitly defined by the "x" eXception.
Cheers.