|
|
||||
|---|---|---|---|---|---|
|
|
||||
|
|
||||
|
|
||||
|
| ||||
|
| ||||
|
|
Friday, February 15, 2013
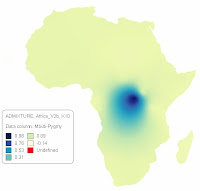
Gradient Maps for African ADMIXTURE components
Here below are gradient maps for my last African ADMIXTURE run, Africa_V2b, courtesy of a demo download of Mapviewer7 . The Kriging method was used for Gridding and 'Grid Z limits' mode was used for color mapping.
UPDATE (02/18/2013) : Below are gradient maps for the first African ADMIXTURE run, Africa_V1, courtesy of a demo download of Mapviewer7 . The same options as above were used both for gridding and color mapping.
Subscribe to:
Post Comments (Atom)






























The bimodal Omotic distribution is a bit surprising. Is there any logical reason why that should be present?
ReplyDeleteIt is because of the high presence of the omotic cluster found in the Sandawe of Tanzania, possibly an ancient rift valley presence that got disrupted by incoming farmers from the west.
DeleteThis test is interesting.
ReplyDeleteI am mostly European with about 5% middle eastern in my DNA from other sites. I am according to the ethohelix with gedmatch 50% Egyptian, 25% Pakistan and 25% Moroccan.
Now with the other DNA tests I have 20+% Italian/French region of the world so maybe it's just a carry over
I don't understand how you could possibly get a Pakistani reference, as I have never used any Pakistanis for any of the allele frequency results generated with ADMIXTURE that I have submitted to GEDmatch.
DeleteIf you can tell me exactly which test you used to come up with those results I can give you a more helpful input.
The only Calculators on GEDmatch that include Pakistani populations are MDLP Ancient Roots K18 and Dodecad V3
ReplyDeleteJohn Olson - Administrator, GEDmatch
Thanks, that's what I thought, perhaps James Wolfe misread Palestinian for Pakistani?
DeleteHello, Thank you for this wonderful tool! My results of DNA testing showed that I am 74% Sub Saharan west African, 1.7% East Asian & Native American and 0.2% Middle East & North African, 18% European . The EthioHelix K10 + Palestinian came up with 89.89% Palestinian. Also on the EthioHelix K10 + Japanese run 74% North Africa and 17% Japanese. The Palestinian percentage was very high. Does this mean there is a Great similarity to the West African genome?
DeleteI wish I knew how to read this data. I can't make any English out of it.
DeleteMy question is do I have a connection with the Ethiopian Jews according to your findings?
ReplyDeleteHello Ethio Helix, I was noticing something interesting with your calculator. It seems to me that when Europeans estimate their ADMIXTURE Proportion using the Ethio Helix K10 Africa Only calculator are divided into two clusters. North-Africa and East-Africa2. However, when Middle Easterners such as myself calculate our ADMIXTURE Proportions we tend to register more of the East-Africa2 and less of the North-Africa component while with Europeans this seems to have the opposite effect.
ReplyDeleteWould this imply that Middle Easterners have a higher affinity to East African populations compared to Europeans?
"Would this imply that Middle Easterners have a higher affinity to East African populations compared to Europeans?"
DeleteYes, I would tend to think that would naturally be the case. The Africa only calculator has no non-African populations used in the analysis and the fst layout of the ADMIXTURE components can be seen here. As you can see the EA2 and NA components are close to each other, but while the NA component registers highest (in terms of frequency) with the Maghrebi populations in the dataset, the EA2 component registers highest with East African populations.
Now what happens if we start appending non-African populations to this dataset, say starting with Middle Easterners (ME), then Europeans , then South Asians and so forth, what happens is that this global genetic cline starts forming and completes itself with the addition of East Asians and Native Americans. As you can see in the cline, MEs are closer to EAs than Europeans are, therefore it is only natural that when the only available populations in the dataset are Africans, MEs would register more EA2 than Europeans.
Now the reason to this affinity is a different ball of wax altogether, we have several lines of evidence that show after the initial OOA migrations, several bidirectional movements have occurred between EA and ME for a very long time. However, older (and even some newer) academia has taken the stance of explaining away this affinity with uni-directional punctuated movements , to be sure the direction only being from ME --> EA. I disagree with this assessment, and in my humble opinion think that it is rooted in simple old-fashioned bigotry against Africans and not reasonable logical deductions.
"Now the reason to this affinity is a different ball of wax altogether, we have several lines of evidence that show after the initial OOA migrations, several bidirectional movements have occurred between EA and ME for a very long time. However, older (and even some newer) academia has taken the stance of explaining away this affinity with uni-directional punctuated movements , to be sure the direction only being from ME --> EA. I disagree with this assessment, and in my humble opinion think that it is rooted in simple old-fashioned bigotry against Africans and not reasonable logical deductions."
DeleteI agree, I think that the discovery of the Basal Eurasian cluster by Lazaridis et al. via the EEF component present in the Stuttgart farmer revealed undeniable African affinities. Which were later confirmed from other analyses such as Tree Mix and other formal software showing gene flow from African populations, be they Yoruba, Hadza, Dinka, etc. to Stuttgart. Which led some people to extrapolate that Basal Eurasian is either an Aboriginal Arabian component, which I personally think is the less convincing hypothesis, or that it may originally be an East African component. Which some think is far fetched, but to me this actually makes sense, since we know of an ancient harvesting community (Natufians) from the Levant being described as and clustering with African populations.
The anthropologists at the time didn't think there was any genetic continuity between the Natufians and the present day populations in the Middle East. Now I'm starting to think they were wrong, and that the Natufians were actually rich in this Basal Eurasian component, which they contributed to Middle Easterners.
This comment has been removed by the author.
DeleteGreetings Ethio Helix,
DeleteYou're spot on regarding the "uni-directional" bias (ME->EA) embedded in academia. It seems to me that the standard narrative is attempting to project that "high civilization" was imported into Africa (via eastern sphere) by so-called "Eurasian" cultural bearers. Western academia latch to one set of facts ("back migration") but suppress the high possibility of several out migrations that fed and nurtured civilizations such as Sumeria. This same line of thinking follows with "Afroasiatic" languages: That this phylum penetrated into Africa via East when in all probability its origins was in Ethiopia or adjacent region on the African continent. Finally, when I brought this question (origins of Afroasiatic language) to my linguistics professor (Dr. Hailu) then as an undergraduate student, he put forth the analogy "that's like taking your right hand and trying to wrap it around your head to touch your right ear," his words exactly. I'd say this equally applies to the uni-directional "stance" that you mentioned. Please so keep up the good work!
This comment has been removed by the author.
ReplyDeleteHello Ethio Helix: what an interesting feature on gedmatch! thank you. I'm quite a layperson here, so I wish there was a 3rd grade level explanation of what I'm looking at. I'm about 2/3 European, but about 1/3 Middle Eastern. I did the French to African Proportion. A lot of small Omotic percentages; 1.5% Hadza; 2% Khoi-San; 1% Eastern Bantu; a few percentages of Nilo-Saharan, including 2.8%; and as expected some very high percentages of North African on all but 2 chromosomes. I guess my main question is, is the French aspect. Is the French % just a general percentage of Northwestern European?
ReplyDeleteI'm not a scientist, so an easy explanation would be very much appreciated! Thank you for this very interesting feature and blog!
ok, now I just did the Palestinian combination, and it's all over the map!
Delete" I guess my main question is, is the French aspect. Is the French % just a general percentage of Northwestern European?"
DeleteYes, or European generally, the calculators are only as good as the populations referenced in the initial ADMIXTURE run, you can see those here, you can also click on the clusters to sort any specific cluster's distribution amongst the referenced populations. In the Africa + French calculator for instance, only the french are referenced amongst Europeans so they behave as a proxy for other European and close by populations, and you can even see some Africans having the French component, does that mean those Africans have actual French Admixture, No, it just means that a portion of their genome that is registered as 'French' has more similarity/Affinity to the referenced French genomes than it does to the other clusters that were found in Africa. You can think of the other clusters in the same vein.
OK, thank you for the explanation. I guess the more Southern African percentages came as a bit of a surprise for me. Very interesting! Thank you.
DeleteThis comment has been removed by the author.
ReplyDeleteMy results for the EthioHelix K10 Africa Only Admixture Proportions were:
ReplyDeleteNilo-Saharan 4.31
East-Africa2 3.87
Mbuti-Pygmy 1.79
East_Africa1 1.02
Khoi-San 2.02
West_Africa 74.94
Hadza 1.18
Biaka-Pygmy 4.28
North-Africa 4.23
Omotic 2.36
However, on Ancestry they were: Ghana 71%, Nigeria 13%, Benin-Togo 5%, Cameroon-Cong and Mali 2% each and Caucasian 6%.
I would like to know if you used the Ancestry reference populations (e.g. Brong or Yoruba), and whether your website can offer more detail on the results?
So I ran the test k10+palestine and I came back 91% palestine. I'm so so so new at this and all the other tests have been saying I'm Baltic or euopean. Cam you please explain how this test is broken down in comparison.
ReplyDeleteThis comment has been removed by the author.
DeleteMine has come back the same it says 91% Palestinian. I know I have ashkanazi but I don't understand where the high percentage of the Palestinian is from. Could someone explain?
ReplyDeleteMy results came back 91% palestinian too and I have Ashkenazi, perhaps it is coming from a distance ancestor ?
ReplyDeleteHi, Ethio Helix
ReplyDeleteI have some questions about the ethiohelix Africa + French. Im a mixed person. Mostly european though. I have some west Asian and american indian. I am about 30% Mediterranean from south Europe. I heard that Sicilian and Spain/ Portuguese are admixed with North African and middle eastern. So I decided to put my raw dna into the ethiohelix Africa + French and it says I'm 81% French and like 16% north african, 1% omotic, 1% hadza, and 1% mbuti-pygmy. I'm confused with breakdown and does that mean my Mediterranean had a lot of North African admixed? What do these African percentages represent?
I recently received my new results from FTDNA and plugged them into the EthioHelix K10 + Palestinian Admixture Proportions. I am a caucasian male in the US. How do I interpret these results?;
ReplyDeleteNilo-Saharan -
East-Africa1 0.48
Mbuti-Pygmy 0.51
Eastern-Bantu -
Khoi-San 1.13
West-Africa -
Hadza 0.97
Biaka-Pygmy 0.69
Palestinian 91.08
Omotic 5.13
Hello, I have questions about my ethiohelix Africa + French results. My ancestry Dna results are 20% Mali, 7% Nigerian, 4% Southeastern Bantu, 1% Benin/Togo. I am 60% European, mostly Iberian, Italy/Greece, then Irish and French. It also have 4% Native American.
ReplyDeleteMy Ethiohelix Africa+French results are French 48.95%, North Africa 15.61%, Eastern Bantu 10.11%, West Africa, 14.18%, Omotic 8.77%, Biaka-Pigmy 2.36%. I was surprised by the North Africa results and the East African results. Is it showing less Euro and the amount of North African because of my Southern European ancestry? Also, I was hoping you could explain the West African, and East African results compared to Ancesry. Being of African American decent, most of the enslaved Africans came from the West. However, I was pleasantly surprised by those results. Does Omotic mean I’m part Ethiopian?
This comment has been removed by the author.
DeleteThis comment has been removed by the author.
DeleteHi Angela D,
DeleteI hope to be able to shed some light on the situation.
Prior to doing the Ethiohelix Africa+French and Ancestry DNA I identified as either Nigerian (Yoruba tribe), West African or Black British.
My Ancestry results were similar to yours with respect to African regions but higher percentages. However, both of my parents and grandparents were born in Nigeria, West Africa and claimed to be West African through and through. My Ethiohelix Africa+French results were again displaying similar regions to yours but French was very low percentage. This reminded me of stories that had been passed down from my dad's side of the family, growing up my dad had always advised that originally although we are West African, our ancestors were nomadic people and had crossed the Sahara from East to West, specifically Ethiopia. I was also advised by my dad that prior to colonisation, African's identified themselves by their tribe rather than by country borders. Tribes also intermarried which explaines the admixtures in West Africa.
In light of the above, although the slaves were taken from West Africa, most West African's have many admixtures from all parts of Africa. On an interested note, having done the Ancestry DNA I discovered that I had many 4th and 5th African American cousins (especially from the Southern States), yet growing up I was told that my family had no slave ancestry.
I hope that the above had helped :)
Population
DeleteNilo-Saharan 3.89
North-Africa -
Mbuti-Pygmy 2.19
Eastern-Bantu 17.34
Khoi-San 1.28
West-Africa 64.63
Hadza 2.90
Biaka-Pygmy 5.56
French 0.38
Omotic 1.81
Can you help me understand? My father is African American and my mother is of European descent. My results from this calculator:
ReplyDeleteNilo-Saharan 0.94
East-Africa1 2.16
Mbuti-Pygmy 1.03
Eastern-Bantu 8.96
Khoi-San 0.63
West-Africa 8.84
Hadza 1.87
Biaka-Pygmy 3.63
Palestinian 67.44
Omotic 4.49
Does this group my European in Palestinian?
DeleteI am mostly European (Blond, blue eyes) but adopted and unknown ancestry so I took this test and my results came back as:
ReplyDeleteNilo-Saharan -
North-Africa 20.76
Mbuti-Pygmy 0.35
Eastern-Bantu -
Khoi-San -
West-Africa -
Hadza 0.75
Biaka-Pygmy -
French 75.91
Omotic 2.24
I was not expecting such a large North African %, as it doesn't match my other tests, but can North African be the same genes as Mediterranean? I do have various (10-24%) Mediterranean results in several GEDMatch tests.
The other noticeable result being Omotic, however in the EuroGenes K36 test it lists Omotic as 0.
As for Hadza, I have no other tests with it listed to compare, but MDLP World-22 does list Pygmy as a result, though again EuroGenes K36 says 0.
Hi! My Ethiohelix results are nearly identical to yours (also blonde with blue eyes). In the near term, I've mostly descended from the Acadian people... very early French colonists in present-day Nova Scotia. They all ended up in Southeast Louisiana in the 18th century where they mixed with many other ethnicities. While America was still colonial, there was a significant population of people from the Canary Islands that mixed a lot with the now Louisianian Acadians. Spain had long colonized the Canary Islands by the time they immigrated to North America, but going further back, it's believed that they'd been settled earlier by Northern Africans as well as fishing tribes elsewhere around the Mediterranean (they were eventually Hispanized by Spain).
DeleteI've traced many ancestors with Spanish names on both sides to the Canary Islands (as most people of Louisiana Acadian descent probably can), but my Ancestry.com results (and that of my mom) shows more North African descent than Spanish descent. I can only suspect it's because most of my Canary Islander ancestors weren't actually genetically Hispanic. Over time, they mixed with all the other groups that migrated to or through New Orleans... lots of Sicilians, Irish, and German, as well as Africans brought through slave trade. For various reasons, there was always a lot more interracial and interethnic mixing in the region than elsewhere in the U.S., and that may explain trace amounts of genetics from other regions of Africa, as well as the overall jumble of European heritage.
French 75.43 Pct
North-Africa 20.79 Pct
Omotic 3.07 Pct
Khoi-San 0.71 Pct
Hello, I'm completely from Africa, but on the Ethiohelix Africa + Japanese test, It gave me about 5% Japanese. I didn't have anything like that in Africa + Palestine (had 0% palestinian) or Africa + French (had 0% French). So does that me I have potential Asian ancestry hidden in there?
ReplyDelete1 French 42.71
ReplyDelete2 West-Africa 16.9
3 North-Africa 16.39
4 Omotic 9.38
5 Eastern-Bantu 5.32
6 Biaka-Pygmy 2.57
7 Mbuti-Pygmy 2.56
8 Nilo-Saharan 1.96
9 Khoi-San 1.69
10 Hadza 0.53
my results and here's my question is there any way to narrow down the tribes or possible tribes we descend from or do we have to take that 300 dollar test?
Palestinian 56.48
ReplyDelete2 Omotic 9.49
3 West-Africa 9.39
4 Eastern-Bantu 7.22
5 Biaka-Pygmy 5.23
6 Nilo-Saharan 3.38
7 Mbuti-Pygmy 2.84
8 East-Africa1 2.32
9 Khoi-San 2.26
10 Hadza 1.39
palestinian results
1 North-Africa 41.4
ReplyDelete2 Japanese 17.83
3 West-Africa 11.87
4 Omotic 10.41
5 Biaka-Pygmy 5.92
6 Nilo-Saharan 4.04
7 East-Africa1 3.08
8 Khoi-San 2.69
9 Mbuti-Pygmy 2.56
10 Hadza 0.
japanese results
my results and here's my question is there any way to narrow down the tribes or possible tribes we descend from or do we have to take that 300 dollar test?
Biggjosh, have you taken a Y-chromosome test? If you should happen to have a close match with an African's Y-DNA haplotype, that would be a good indication of the origins of your paternal lineage. The test by African Ancestry that I think you're referring to, isn't considered very valid, because they use a tiny number of markers, so you'll match people in many ethnic groups and countries. The more markers you have tested, the higher the accuracy and reliability of the match.
DeleteMy research project on the A00 haplogroup has resulted in our collecting 900 samples from Cameroon. These have all been tested to at least 18 markers, which isn't ideal, but is much better than what African Ancestry is doing. I also have a large collection of Y-DNA haplotypes from other countries in Africa, which also have had a decent number of markers tested.
I'd like to invite anyone interested to let me search this data for matches with your Y-DNA, for a modest fee, much less than $300. If you haven't had a Y-DNA test yet, you can get one from YSEQ at a very reasonable cost, the lowest currrently available. If you match one of our samples from Cameroon, you may get it upgraded to 37 or more markers, so your sample can be compared with it more exactly, and we can estimate your TMRCA.
Please, anyone who is interested, contact DNA RootSearch on Facebook or at DNArootsearch.com (This should be on a new web page in the future, but for now, that's where you can get in touch.
Oh, I should have mentioned that all this data includes the ethnicity (tribe) and location of the person whose DNA was tested, but our own Cameroonian data includes much more detail, such as the languages spoken, the village where it was collected, and the birthplaces of the sample donor, his father, and grandfather.
DeleteI tested for a large percentage of North African (equivalent to Arabic/Middle East marker) on the Japanese calculator and Palestinian (also a Middle East marker) on the Palestinian calculator which is spot on since I have a significant Iberian/South European/Middle East DNA marker on almost every calculator and DNA test. But here is lies my question. I also showed a big Japanese DNA marker (21%) on the Japanese calculator. I do have small Siberian/Asian/Amerindian DNA results on other tests & calculators--which minimally supports my known Native American ancestry. Can this large Japanese DNA % be interpreted to reliably support the other Amerindian Siberian/Asian markers AND to strengthen the DNA evidence supporting these "Asian" markers?
ReplyDelete