There
are two ways of choosing a K value for any given dataset that one
wishes to perform an ADMIXTURE run on, one is to throw a dart at a
random set of numbers and hope it works out for the very best, the other
is to run ADMIXTURE at different K's while computing a cross
validation error for each of the K values using the --cv flag, I did
this with the studentized global dataset that I discussed earlier in this post. The Cross Validation error values for K 1-14 for that particular dataset can be
seen in the graphs below,
close up :
While
the CV-Error values do not start flattening out until about K=10, the
CV error values do not start inflecting until K=13, meaning K=13 is
the appropriate choice for this dataset.
Cross
Validation can take a considerably long time to run, as each
consecutive K has to be evaluated along with its error
separately, unless one has access to a very fast machine off-course.
As
a reference, the Bash shell code to run Cross Validation in ADMIXTURE
for up-to K=14 is:
for
K in 1 2 3 4 5 6 7 8 9 10 11 12 13 14; \
do
./admixture32 -j2 --cv=14 “filename.bed” $K | tee log${K}.out;
done
where
CV error values will be recorded in the .out files for each K.
Peaking
populations for each cluster for K =2-13
K=2
Cluster1:
pygmy,mbutipygmy,sotho/tswana,biakapygmy,fang
Cluster2:
chinese-americans,tujia,miao,hezhen,han
East Asians and Africans split, with West Asians and Europeans
belonging to 1/3 African and 2/3 East Asian, the reverse is seen with Ethiopians,
2/3 African and 1/3 East Asian.
K=3
Cluster1:
sardinian,basque,tuscans,italian,spaniards
Cluster2:
pygmy,mbutipygmy,sotho/tswana,biakapygmy,bantusouthafrica
Cluster3:
she,chinese-americans,han,singapore-chinese,chinese
K=4
Cluster1:
sardinian,basque,tuscans,italian,cypriots
Cluster2:
pygmy,mbutipygmy,sotho/tswana,biakapygmy,bantusouthafrica
Cluster3:
colombian,karitiana,surui,pima,totonac
Cluster4:
she,han,singapore-chinese,chinese,miao
K=5
Cluster1:
she,han,chinese-americans,chinese,singapore-chinese
Cluster2:
surui,karitiana,colombian,pima,totonac
Cluster3:
sardinian,basque,spaniards,italian,tuscans
Cluster4:
pygmy,mbutipygmy,biakapygmy,bantusouthafrica,sotho/tswana
Cluster5:
papuan,irula,tn-dalit,ap-mala,malayan
K=6
Cluster1:
papuan,melanesian,tongan,samoan,paniya
Cluster2:
pygmy,mbutipygmy,biakapygmy,bantusouthafrica,sotho/tswana
Cluster3:
karitiana,colombian,surui,pima,totonac
Cluster4:
she,han,chinese-americans,singapore-chinese,chinese
Cluster5:
sardinian,basque,spaniards,italian,tuscans
Cluster6:
irula,tn-dalit,ap-madiga,ap-mala,north-kannadi
K=7
Cluster1:
sardinian,basque,spaniards,italian,tuscans
Cluster2:
dogon,yoruba,bambaran,hausa,igbo
Cluster3:
irula,tn-dalit,ap-mala,ap-madiga,north-kannadi
Cluster4:
san-nb,san,!kung,pygmy,mbutipygmy
Cluster5:
papuan,melanesian,tongan,samoan,paniya
Cluster6:
colombian,surui,karitiana,pima,totonac
Cluster7:
she,han,chinese-americans,singapore-chinese,chinese
K=8
Cluster1:
dogon,yoruba,bambaran,hausa,igbo
Cluster2:
irula,tn-dalit,ap-mala,ap-madiga,north-kannadi
Cluster3:
papuan,melanesian,tongan,samoan,paniya
Cluster4:
koryaks,nganassans,chukchis,evenkis,yakut
Cluster5:
dai,vietnamese,singapore-chinese,she,han
Cluster6:
sardinian,basque,spaniards,italian,tuscans
Cluster7:
san-nb,san,!kung,pygmy,mbutipygmy
Cluster8:
surui,karitiana,colombian,pima,totonac
K=9
Cluster1:
papuan,melanesian,tongan,samoan,paniya
Cluster2:
iban,samoan,tongan,singapore-malay,dai
Cluster3:
japanese,hezhen,han-nchina,xibo,beijing-chinese
Cluster4:
sardinian,basque,spaniards,italian,tuscans
Cluster5:
san-nb,san,!kung,pygmy,mbutipygmy
Cluster6:
dogon,yoruba,bambaran,hausa,igbo
Cluster7:
surui,karitiana,colombian,pima,totonac
Cluster8:
irula,tn-dalit,ap-mala,ap-madiga,north-kannadi
Cluster9:
koryaks,chukchis,nganassans,east-greenlanders,kets
K=10
Cluster1:
saudis,bedouin,yemen-jews,samaritians,tunisia
Cluster2:
papuan,melanesian,tongan,samoan,paniya
Cluster3:
dai,vietnamese,iban,singapore-chinese,she
Cluster4:
hadza,maasai,ethiopians,ethiopian-jews,bulala
Cluster5:
irula,tn-dalit,ap-madiga,ap-mala,north-kannadi
Cluster6:
surui,karitiana,colombian,pima,totonac
Cluster7:
koryaks,nganassans,chukchis,evenkis,yakut
Cluster8:
dogon,yoruba,brong,igbo,bambaran
Cluster9:
san-nb,san,!kung,pygmy,mbutipygmy
Cluster10:
lithuanians,belorussian,orcadian,n-european,utahn-whites
West Asian component splits into 2 components; North European and
Middle East & North African (MENA). An East African component that was previously concealed by the West Asian and African components forms. The previous South East Asian component
disappears.
K=11
Cluster1:
dai,vietnamese,singapore-chinese,she,han
Cluster2:
koryaks,nganassans,chukchis,evenkis,yakut
Cluster3:
surui,karitiana,colombian,pima,totonac
Cluster4:
tunisia,bedouin,saudis,sahara-occ,yemen-jews
Cluster5:
dogon,yoruba,brong,igbo,bambaran
Cluster6:
lithuanians,belorussian,orcadian,n-european,utahn-whites
Cluster7:
papuan,melanesian,tongan,samoan,paniya
Cluster8:
san-nb,san,!kung,pygmy,mbutipygmy
Cluster9:
irula,malayan,tn-dalit,ap-mala,ap-madiga
Cluster10:
hadza,maasai,ethiopians,sandawe,bulala
Cluster11:
kalash,brahui,balochi,makrani,georgians
K=12
Cluster1:
surui,karitiana,colombian,pima,totonac
Cluster2:
lithuanians,belorussian,orcadian,n-european,utahn-whites
Cluster3:
san-nb,san,!kung,pygmy,mbutipygmy
Cluster4:
iban,samoan,tongan,singapore-malay,cambodian
Cluster5:
bedouin,saudis,yemen-jews,samaritians,tunisia
Cluster6:
papuan,melanesian,tongan,samoan,paniya
Cluster7:
japanese,beijing-chinese,han-nchina,chinese-americans,xibo
Cluster8:
koryaks,chukchis,east-greenlanders,west-greenlanders,kets
Cluster9:
irula,tn-dalit,ap-madiga,ap-mala,north-kannadi
Cluster10:
dogon,yoruba,brong,igbo,bambaran
Cluster11:
nganassans,evenkis,yakut,dolgans,kets
Cluster12:
hadza,maasai,ethiopians,ethiopian-jews,bulala
Central Asian component disappears, a second Siberian component is
formed, the S. East Asian component reappears.
K=13
Cluster1:
san-nb,san,!kung,xhosa,bantusouthafrica
Cluster2:
surui,karitiana,colombian,pima,totonac
Cluster3:
papuan,melanesian,tongan,samoan,paniya
Cluster4:
japanese,han-nchina,beijing-chinese,xibo,hezhen
Cluster5:
hadza,maasai,ethiopians,sandawe,bulala
Cluster6:
lithuanians,belorussian,orcadian,n-european,utahn-whites
Cluster7:
koryaks,chukchis,nganassans,evenkis,east-greenlanders
Cluster8:
tunisia,bedouin,saudis,yemen-jews,sahara-occ
Cluster9:
kalash,brahui,balochi,makrani,georgians
Cluster10:
pygmy,mbutipygmy,biakapygmy,alur,fang
Cluster11:
irula,malayan,tn-dalit,ap-mala,ap-madiga
Cluster12:
dogon,yoruba,brong,bambaran,igbo
Cluster13:
iban,samoan,tongan,singapore-malay,dai
Central Asian Component reappears, a new Pygmy component is formed,
second Siberian component disappears.
Fst
for K=13.
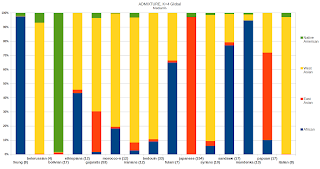
UPDATE: Median cluster % for all populations, K13.
| ADMIXTURE, Global K13 | N | San | N. American | Oceanian | E. Asian | E. African | N. European | Siberian | MENA | Central Asian | Pygmy | S. Asian | W. African | S.E. Asian | |
| !kung | 8 | 78% | 0% | 0% | 0% | 2% | 0% | 0% | 0% | 0% | 2% | 0% | 16% | 0% | |
| adygei | 11 | 0% | 1% | 0% | 3% | 0% | 32% | 3% | 20% | 42% | 0% | 1% | 0% | 0% | |
| african-americans | 37 | 2% | 1% | 0% | 0% | 1% | 13% | 0% | 1% | 3% | 3% | 0% | 72% | 0% | |
| algeria | 12 | 0% | 0% | 0% | 0% | 5% | 22% | 1% | 48% | 5% | 0% | 3% | 13% | 0% | |
| altaians | 8 | 0% | 2% | 0% | 37% | 0% | 12% | 31% | 0% | 12% | 0% | 0% | 0% | 0% | |
| alur | 7 | 0% | 0% | 0% | 0% | 34% | 0% | 0% | 0% | 0% | 17% | 0% | 50% | 0% | |
| ap-brahmin | 14 | 0% | 1% | 2% | 1% | 0% | 8% | 2% | 1% | 36% | 0% | 48% | 0% | 2% | |
| ap-madiga | 5 | 0% | 0% | 2% | 2% | 0% | 0% | 0% | 0% | 24% | 0% | 66% | 0% | 5% | |
| ap-mala | 8 | 0% | 0% | 2% | 2% | 0% | 0% | 0% | 0% | 22% | 0% | 67% | 0% | 5% | |
| armenians | 11 | 0% | 0% | 0% | 0% | 0% | 19% | 0% | 34% | 43% | 0% | 2% | 0% | 0% | |
| armenians-b | 3 | 0% | 0% | 1% | 0% | 0% | 48% | 4% | 17% | 26% | 0% | 1% | 0% | 0% | |
| ashkenazy-jews | 15 | 0% | 0% | 0% | 1% | 0% | 37% | 0% | 34% | 24% | 0% | 1% | 0% | 0% | |
| azerbaijan-jews | 6 | 0% | 1% | 0% | 0% | 0% | 15% | 0% | 37% | 44% | 0% | 0% | 0% | 1% | |
| balochi | 18 | 0% | 1% | 0% | 1% | 0% | 7% | 1% | 13% | 53% | 0% | 20% | 0% | 0% | |
| bambaran | 14 | 3% | 1% | 0% | 0% | 1% | 0% | 0% | 1% | 0% | 1% | 0% | 91% | 0% | |
| bamoun | 10 | 3% | 0% | 0% | 0% | 4% | 0% | 0% | 0% | 0% | 7% | 0% | 85% | 0% | |
| bantukenya | 5 | 3% | 0% | 0% | 0% | 20% | 0% | 0% | 2% | 0% | 5% | 0% | 67% | 0% | |
| bantusouthafrica | 3 | 24% | 0% | 0% | 1% | 6% | 0% | 0% | 0% | 0% | 4% | 0% | 65% | 0% | |
| basque | 24 | 0% | 0% | 1% | 0% | 0% | 75% | 0% | 16% | 6% | 0% | 1% | 0% | 0% | |
| bedouin | 33 | 0% | 0% | 0% | 0% | 3% | 0% | 0% | 65% | 27% | 0% | 0% | 2% | 0% | |
| beijing-chinese | 91 | 0% | 0% | 0% | 68% | 0% | 0% | 2% | 0% | 0% | 0% | 0% | 0% | 28% | |
| belorussian | 4 | 0% | 1% | 1% | 0% | 0% | 77% | 4% | 3% | 15% | 0% | 1% | 0% | 0% | |
| biakapygmy | 12 | 17% | 0% | 0% | 0% | 1% | 0% | 0% | 0% | 0% | 33% | 0% | 45% | 0% | |
| bnei-menashe-jews | 4 | 0% | 0% | 2% | 1% | 0% | 7% | 0% | 16% | 34% | 0% | 34% | 0% | 3% | |
| bolivian | 17 | 0% | 95% | 0% | 1% | 0% | 1% | 3% | 0% | 0% | 0% | 0% | 0% | 0% | |
| brahui | 18 | 0% | 1% | 0% | 0% | 0% | 8% | 1% | 13% | 55% | 0% | 20% | 0% | 0% | |
| brong | 4 | 4% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 1% | 3% | 0% | 91% | 0% | |
| bulala | 12 | 0% | 0% | 0% | 0% | 38% | 0% | 0% | 3% | 0% | 0% | 0% | 57% | 0% | |
| burusho | 17 | 0% | 2% | 1% | 7% | 0% | 13% | 4% | 2% | 41% | 0% | 27% | 0% | 2% | |
| buryat | 16 | 0% | 0% | 1% | 49% | 0% | 5% | 38% | 1% | 5% | 0% | 0% | 0% | 1% | |
| buryats | 13 | 0% | 0% | 1% | 47% | 0% | 5% | 38% | 0% | 5% | 0% | 1% | 0% | 0% | |
| cambodian | 5 | 0% | 0% | 1% | 31% | 0% | 0% | 0% | 0% | 1% | 0% | 11% | 0% | 57% | |
| chinese | 5 | 0% | 0% | 0% | 60% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 38% | |
| chinese-americans | 73 | 0% | 0% | 0% | 63% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 36% | |
| chukchis | 11 | 0% | 17% | 0% | 0% | 0% | 0% | 80% | 0% | 0% | 0% | 0% | 0% | 2% | |
| chuvashs | 12 | 0% | 2% | 0% | 6% | 0% | 54% | 19% | 1% | 15% | 0% | 2% | 0% | 0% | |
| cochin-jews | 4 | 0% | 2% | 2% | 0% | 1% | 5% | 2% | 8% | 34% | 0% | 46% | 0% | 1% | |
| colombian | 6 | 0% | 100% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | |
| cypriots | 7 | 0% | 0% | 1% | 1% | 0% | 29% | 0% | 39% | 30% | 0% | 0% | 0% | 0% | |
| dai | 6 | 0% | 0% | 0% | 36% | 0% | 0% | 0% | 0% | 0% | 0% | 3% | 0% | 62% | |
| daur | 8 | 0% | 1% | 1% | 63% | 0% | 1% | 25% | 0% | 1% | 0% | 0% | 0% | 8% | |
| dogon | 24 | 1% | 0% | 0% | 0% | 0% | 0% | 0% | 1% | 0% | 0% | 0% | 94% | 0% | |
| dolgans | 5 | 0% | 0% | 0% | 28% | 0% | 10% | 56% | 0% | 3% | 0% | 2% | 0% | 0% | |
| druze | 30 | 0% | 0% | 0% | 0% | 0% | 17% | 0% | 42% | 38% | 0% | 0% | 0% | 0% | |
| east-greenlanders | 6 | 0% | 35% | 0% | 0% | 0% | 4% | 60% | 0% | 0% | 0% | 0% | 0% | 0% | |
| egypt | 12 | 0% | 0% | 0% | 0% | 7% | 11% | 0% | 47% | 24% | 0% | 0% | 7% | 0% | |
| egyptans | 7 | 0% | 0% | 0% | 0% | 8% | 10% | 0% | 49% | 23% | 0% | 0% | 7% | 0% | |
| ethiopian-jews | 12 | 1% | 0% | 1% | 0% | 37% | 0% | 0% | 38% | 8% | 0% | 0% | 11% | 0% | |
| ethiopians | 12 | 1% | 0% | 0% | 1% | 36% | 0% | 1% | 39% | 7% | 0% | 0% | 11% | 0% | |
| evenkis | 11 | 0% | 0% | 0% | 34% | 0% | 3% | 61% | 0% | 2% | 0% | 0% | 0% | 0% | |
| fang | 7 | 6% | 0% | 0% | 0% | 5% | 0% | 0% | 0% | 0% | 7% | 0% | 80% | 0% | |
| french | 22 | 0% | 1% | 0% | 0% | 0% | 70% | 0% | 14% | 12% | 0% | 1% | 0% | 0% | |
| fulani | 7 | 2% | 0% | 0% | 1% | 5% | 7% | 1% | 25% | 0% | 0% | 2% | 58% | 0% | |
| georgia-jews | 4 | 0% | 0% | 0% | 1% | 0% | 16% | 0% | 37% | 43% | 0% | 0% | 0% | 0% | |
| georgians | 17 | 0% | 0% | 0% | 0% | 0% | 23% | 0% | 28% | 46% | 0% | 0% | 0% | 0% | |
| gujaratis | 53 | 0% | 1% | 1% | 1% | 0% | 2% | 0% | 0% | 37% | 0% | 55% | 0% | 2% | |
| gujaratis-b | 14 | 0% | 2% | 1% | 0% | 0% | 13% | 2% | 0% | 40% | 0% | 40% | 0% | 1% | |
| hadza | 11 | 19% | 0% | 0% | 0% | 80% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | |
| han | 24 | 0% | 0% | 0% | 60% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 39% | |
| han-nchina | 6 | 0% | 0% | 0% | 68% | 0% | 0% | 4% | 0% | 2% | 0% | 0% | 0% | 24% | |
| hausa | 9 | 1% | 0% | 0% | 0% | 2% | 0% | 0% | 0% | 0% | 3% | 0% | 90% | 0% | |
| hazara | 16 | 0% | 1% | 0% | 31% | 0% | 14% | 16% | 6% | 23% | 0% | 8% | 0% | 4% | |
| hema | 11 | 3% | 0% | 1% | 0% | 31% | 0% | 1% | 10% | 2% | 4% | 0% | 46% | 0% | |
| hezhen | 4 | 0% | 1% | 0% | 66% | 0% | 0% | 28% | 0% | 0% | 0% | 0% | 0% | 6% | |
| hungarians | 9 | 0% | 2% | 0% | 0% | 0% | 69% | 2% | 10% | 15% | 0% | 1% | 0% | 0% | |
| iban | 15 | 0% | 0% | 2% | 11% | 0% | 0% | 2% | 0% | 0% | 0% | 7% | 0% | 77% | |
| igbo | 10 | 3% | 0% | 0% | 0% | 1% | 0% | 0% | 0% | 0% | 2% | 0% | 90% | 0% | |
| iranian-jews | 4 | 0% | 0% | 0% | 1% | 0% | 12% | 1% | 39% | 44% | 0% | 2% | 0% | 0% | |
| iranians | 12 | 0% | 1% | 1% | 0% | 0% | 16% | 1% | 28% | 45% | 1% | 7% | 1% | 0% | |
| iraq-jews | 8 | 0% | 0% | 1% | 0% | 0% | 14% | 0% | 41% | 40% | 1% | 1% | 0% | 1% | |
| irula | 24 | 0% | 0% | 0% | 0% | 0% | 1% | 0% | 2% | 1% | 0% | 89% | 0% | 0% | |
| italian | 8 | 0% | 0% | 1% | 0% | 0% | 60% | 0% | 23% | 14% | 0% | 0% | 0% | 1% | |
| japanese | 154 | 0% | 0% | 1% | 91% | 0% | 0% | 1% | 0% | 0% | 0% | 0% | 0% | 6% | |
| jordanians | 14 | 1% | 0% | 0% | 0% | 3% | 16% | 1% | 42% | 33% | 0% | 1% | 3% | 1% | |
| kaba | 9 | 2% | 0% | 0% | 1% | 10% | 0% | 0% | 0% | 0% | 4% | 0% | 80% | 0% | |
| kalash | 16 | 0% | 2% | 1% | 0% | 0% | 10% | 3% | 0% | 65% | 0% | 16% | 0% | 2% | |
| karitiana | 14 | 0% | 100% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | |
| kets | 2 | 0% | 5% | 0% | 13% | 0% | 19% | 54% | 0% | 8% | 0% | 1% | 0% | 0% | |
| khmer-cambodian | 3 | 0% | 0% | 3% | 27% | 0% | 0% | 0% | 0% | 0% | 0% | 13% | 0% | 55% | |
| kongo | 5 | 3% | 0% | 0% | 0% | 5% | 0% | 0% | 0% | 0% | 6% | 0% | 83% | 0% | |
| koryaks | 13 | 0% | 7% | 0% | 0% | 0% | 0% | 93% | 0% | 0% | 0% | 0% | 0% | 0% | |
| kurd | 16 | 0% | 1% | 1% | 0% | 0% | 19% | 0% | 29% | 46% | 0% | 3% | 0% | 0% | |
| kyrgyzstani | 15 | 0% | 1% | 0% | 40% | 0% | 13% | 24% | 3% | 12% | 0% | 2% | 0% | 3% | |
| lahu | 5 | 0% | 0% | 1% | 42% | 0% | 0% | 1% | 0% | 0% | 0% | 3% | 0% | 52% | |
| lebanese | 3 | 0% | 1% | 2% | 0% | 1% | 20% | 0% | 40% | 33% | 0% | 2% | 2% | 0% | |
| lezgins | 13 | 0% | 2% | 0% | 0% | 0% | 32% | 2% | 16% | 45% | 0% | 1% | 0% | 0% | |
| libya | 9 | 0% | 1% | 1% | 0% | 7% | 17% | 0% | 50% | 10% | 0% | 2% | 9% | 0% | |
| lithuanians | 6 | 0% | 1% | 0% | 0% | 0% | 80% | 2% | 0% | 12% | 0% | 3% | 0% | 0% | |
| luhya | 73 | 2% | 0% | 0% | 0% | 22% | 0% | 0% | 0% | 0% | 6% | 0% | 67% | 0% | |
| maasai | 100 | 2% | 0% | 0% | 0% | 55% | 0% | 0% | 14% | 0% | 1% | 0% | 24% | 0% | |
| mada | 8 | 0% | 1% | 0% | 0% | 22% | 0% | 0% | 0% | 0% | 3% | 0% | 73% | 0% | |
| makrani | 19 | 0% | 1% | 0% | 0% | 0% | 7% | 0% | 15% | 54% | 0% | 18% | 3% | 0% | |
| malayan | 2 | 0% | 1% | 5% | 3% | 0% | 1% | 2% | 0% | 12% | 1% | 70% | 0% | 6% | |
| mandenka | 13 | 3% | 0% | 0% | 0% | 2% | 0% | 0% | 3% | 0% | 1% | 0% | 88% | 0% | |
| maya | 12 | 0% | 86% | 0% | 1% | 0% | 3% | 3% | 2% | 1% | 0% | 0% | 0% | 0% | |
| mbutipygmy | 13 | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 100% | 0% | 0% | 0% | |
| melanesian | 7 | 0% | 0% | 74% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 25% | |
| mexicans | 38 | 0% | 44% | 0% | 1% | 0% | 27% | 2% | 12% | 6% | 0% | 1% | 3% | 0% | |
| miao | 6 | 0% | 0% | 0% | 56% | 0% | 0% | 1% | 0% | 0% | 0% | 0% | 0% | 42% | |
| mongola | 6 | 0% | 1% | 0% | 64% | 0% | 4% | 14% | 1% | 1% | 0% | 0% | 0% | 13% | |
| mongolians | 8 | 0% | 2% | 1% | 46% | 0% | 10% | 30% | 2% | 7% | 0% | 0% | 0% | 2% | |
| moroccans | 5 | 1% | 0% | 0% | 0% | 3% | 18% | 1% | 54% | 0% | 1% | 3% | 15% | 0% | |
| morocco-jews | 7 | 0% | 0% | 0% | 0% | 1% | 32% | 0% | 39% | 23% | 0% | 1% | 2% | 1% | |
| morocco-n | 12 | 0% | 1% | 0% | 0% | 3% | 27% | 0% | 49% | 1% | 0% | 4% | 12% | 0% | |
| morocco-s | 13 | 0% | 0% | 0% | 0% | 5% | 18% | 0% | 50% | 0% | 1% | 3% | 16% | 0% | |
| mozabite | 21 | 0% | 0% | 0% | 0% | 3% | 20% | 0% | 53% | 0% | 0% | 4% | 16% | 0% | |
| n-european | 14 | 0% | 1% | 0% | 0% | 0% | 74% | 1% | 8% | 13% | 0% | 0% | 0% | 0% | |
| naxi | 5 | 0% | 0% | 1% | 63% | 0% | 0% | 6% | 0% | 0% | 0% | 4% | 0% | 26% | |
| nepalese | 17 | 0% | 1% | 1% | 7% | 0% | 11% | 3% | 0% | 35% | 0% | 35% | 0% | 4% | |
| nganassans | 15 | 0% | 0% | 0% | 11% | 0% | 0% | 88% | 0% | 0% | 0% | 0% | 0% | 0% | |
| nguni | 4 | 18% | 0% | 1% | 0% | 6% | 0% | 0% | 0% | 0% | 4% | 0% | 71% | 0% | |
| north-kannadi | 6 | 0% | 0% | 3% | 3% | 0% | 0% | 0% | 0% | 23% | 0% | 65% | 0% | 3% | |
| orcadian | 9 | 0% | 1% | 0% | 0% | 0% | 75% | 2% | 7% | 14% | 0% | 0% | 0% | 0% | |
| oroqen | 7 | 0% | 0% | 0% | 52% | 0% | 0% | 40% | 0% | 0% | 0% | 0% | 0% | 5% | |
| palestinian | 27 | 0% | 1% | 1% | 0% | 3% | 14% | 0% | 46% | 32% | 0% | 1% | 2% | 0% | |
| paniya | 4 | 0% | 0% | 13% | 16% | 0% | 0% | 1% | 0% | 0% | 1% | 14% | 1% | 48% | |
| papuan | 17 | 0% | 0% | 100% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | |
| pathan | 14 | 0% | 2% | 0% | 1% | 0% | 17% | 1% | 6% | 44% | 0% | 26% | 0% | 1% | |
| pedi | 8 | 18% | 0% | 0% | 0% | 5% | 0% | 0% | 0% | 1% | 4% | 0% | 71% | 0% | |
| pima | 11 | 0% | 95% | 0% | 0% | 0% | 0% | 5% | 0% | 0% | 0% | 0% | 0% | 0% | |
| punjabi-arain | 15 | 0% | 2% | 1% | 0% | 0% | 10% | 1% | 4% | 45% | 0% | 34% | 0% | 0% | |
| pygmy | 17 | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 100% | 0% | 0% | 0% | |
| romanians | 9 | 0% | 0% | 0% | 0% | 0% | 55% | 3% | 19% | 19% | 0% | 0% | 0% | 0% | |
| russian | 20 | 0% | 2% | 0% | 0% | 0% | 70% | 9% | 1% | 14% | 0% | 2% | 0% | 1% | |
| sahara-occ | 10 | 0% | 0% | 0% | 0% | 6% | 16% | 1% | 57% | 0% | 0% | 3% | 15% | 0% | |
| sakilli | 4 | 0% | 0% | 3% | 3% | 0% | 1% | 0% | 0% | 25% | 0% | 64% | 0% | 2% | |
| samaritians | 3 | 1% | 0% | 2% | 0% | 0% | 11% | 0% | 49% | 35% | 0% | 1% | 0% | 0% | |
| samoan | 11 | 0% | 0% | 25% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 74% | |
| san | 24 | 88% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | |
| san-nb | 12 | 100% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | |
| sandawe | 17 | 12% | 1% | 0% | 0% | 38% | 0% | 0% | 13% | 1% | 5% | 0% | 29% | 0% | |
| sardinian | 22 | 0% | 0% | 0% | 0% | 0% | 59% | 0% | 35% | 4% | 0% | 0% | 0% | 0% | |
| saudis | 15 | 0% | 0% | 0% | 0% | 4% | 0% | 0% | 63% | 30% | 0% | 0% | 0% | 0% | |
| selkups | 7 | 0% | 5% | 0% | 9% | 0% | 26% | 47% | 0% | 10% | 0% | 1% | 0% | 0% | |
| sephardic-jews | 13 | 0% | 0% | 0% | 0% | 0% | 33% | 0% | 37% | 26% | 0% | 1% | 0% | 0% | |
| she | 9 | 0% | 0% | 0% | 59% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 40% | |
| sindhi | 15 | 0% | 2% | 1% | 0% | 0% | 11% | 1% | 5% | 44% | 0% | 35% | 0% | 0% | |
| singapore-chinese | 70 | 0% | 0% | 0% | 60% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 40% | |
| singapore-indians | 53 | 0% | 1% | 2% | 1% | 0% | 2% | 1% | 1% | 32% | 0% | 54% | 0% | 3% | |
| singapore-malay | 59 | 0% | 1% | 4% | 15% | 0% | 0% | 1% | 0% | 1% | 0% | 10% | 0% | 65% | |
| slovenian | 17 | 0% | 1% | 0% | 0% | 0% | 70% | 2% | 9% | 15% | 0% | 1% | 0% | 0% | |
| sotho/tswana | 5 | 25% | 0% | 0% | 0% | 3% | 0% | 0% | 0% | 0% | 4% | 0% | 67% | 0% | |
| spaniards | 5 | 0% | 0% | 0% | 0% | 0% | 68% | 1% | 19% | 10% | 0% | 0% | 1% | 1% | |
| stalskoe | 5 | 0% | 2% | 0% | 2% | 0% | 34% | 3% | 16% | 39% | 0% | 2% | 0% | 0% | |
| surui | 7 | 0% | 100% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | |
| syrians | 10 | 0% | 1% | 0% | 0% | 1% | 16% | 0% | 40% | 35% | 0% | 3% | 2% | 0% | |
| thai | 17 | 0% | 1% | 2% | 15% | 0% | 1% | 2% | 1% | 3% | 0% | 16% | 0% | 57% | |
| tn-brahmin | 9 | 0% | 2% | 2% | 0% | 0% | 8% | 2% | 0% | 36% | 0% | 48% | 0% | 1% | |
| tn-dalit | 7 | 0% | 0% | 3% | 0% | 0% | 0% | 1% | 0% | 23% | 0% | 67% | 0% | 5% | |
| tongan | 11 | 0% | 0% | 30% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 70% | |
| totonac | 15 | 0% | 91% | 0% | 1% | 0% | 3% | 5% | 0% | 0% | 0% | 0% | 0% | 0% | |
| tu | 7 | 0% | 1% | 1% | 63% | 0% | 3% | 8% | 1% | 3% | 0% | 1% | 0% | 18% | |
| tujia | 5 | 0% | 0% | 0% | 62% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 36% | |
| tunisia | 11 | 0% | 0% | 0% | 0% | 1% | 20% | 0% | 59% | 0% | 0% | 4% | 13% | 0% | |
| turks | 13 | 0% | 1% | 0% | 4% | 0% | 26% | 3% | 28% | 35% | 0% | 2% | 0% | 0% | |
| tuscans | 79 | 0% | 0% | 0% | 0% | 0% | 53% | 0% | 26% | 18% | 0% | 0% | 0% | 0% | |
| tuvinians | 11 | 0% | 1% | 1% | 41% | 0% | 9% | 40% | 0% | 6% | 0% | 0% | 0% | 1% | |
| urkarah | 11 | 0% | 2% | 0% | 0% | 0% | 36% | 2% | 11% | 45% | 0% | 0% | 0% | 0% | |
| utahn-whites | 72 | 0% | 1% | 0% | 0% | 0% | 75% | 1% | 7% | 12% | 0% | 1% | 0% | 0% | |
| uygur | 7 | 0% | 2% | 0% | 29% | 0% | 17% | 12% | 5% | 22% | 0% | 7% | 0% | 6% | |
| uzbekistan-jews | 2 | 0% | 1% | 1% | 0% | 0% | 18% | 1% | 35% | 42% | 0% | 2% | 0% | 1% | |
| uzbeks | 10 | 0% | 1% | 0% | 27% | 0% | 21% | 17% | 6% | 20% | 0% | 6% | 0% | 1% | |
| vietnamese | 4 | 0% | 0% | 1% | 42% | 0% | 0% | 0% | 0% | 0% | 0% | 4% | 0% | 52% | |
| west-greenlanders | 8 | 0% | 26% | 0% | 0% | 0% | 23% | 45% | 1% | 2% | 0% | 2% | 0% | 0% | |
| xhosa | 3 | 27% | 0% | 0% | 0% | 7% | 0% | 0% | 1% | 0% | 2% | 0% | 61% | 0% | |
| xibo | 6 | 0% | 0% | 1% | 67% | 0% | 1% | 15% | 0% | 2% | 0% | 0% | 0% | 13% | |
| yakut | 18 | 0% | 0% | 1% | 37% | 0% | 3% | 53% | 1% | 4% | 0% | 0% | 0% | 0% | |
| yemen-jews | 12 | 0% | 0% | 1% | 0% | 4% | 3% | 0% | 58% | 31% | 0% | 1% | 0% | 0% | |
| yemenese | 7 | 1% | 0% | 1% | 1% | 5% | 3% | 1% | 42% | 28% | 1% | 3% | 7% | 1% | |
| yi | 6 | 0% | 0% | 1% | 62% | 0% | 0% | 7% | 0% | 0% | 0% | 3% | 0% | 26% | |
| yoruba | 92 | 2% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 2% | 0% | 93% | 0% | |
| yukaghirs | 6 | 0% | 0% | 0% | 16% | 0% | 31% | 42% | 0% | 6% | 0% | 1% | 0% | 0% |
All results can be downloaded here: ADMIXTURE_K1-14.tar.gz
which contains:
PLINK formatted *.bed, *.bim, *.fam files
*.txt file with complete list of samples
K folders containing:
*.P and *.Q ADMIXTURE output files
log file, with Fst distances and CV errors
Processed Output folder containing:
Median Cluster %
Average Cluster %
Standard Deviations
Cluster Key: Top five populations in each cluster
list of Unique Populations
GNU OCTAVE variable loading file, *.mat
which contains:
PLINK formatted *.bed, *.bim, *.fam files
*.txt file with complete list of samples
K folders containing:
*.P and *.Q ADMIXTURE output files
log file, with Fst distances and CV errors
Processed Output folder containing:
Median Cluster %
Average Cluster %
Standard Deviations
Cluster Key: Top five populations in each cluster
list of Unique Populations
GNU OCTAVE variable loading file, *.mat



























Why do mandenka have MENA component? Also, many of the North African have significant West African component? this is not seen in other analysis of this type except maybe for some of the South Morroco.
ReplyDeleteI am supposing you are talking about the cross validated K13 results correct ?
DeleteThe Mandenka had the MENA component at 2.83%, other West Africans like the Dogon had it at 1.19%. The MENA component is the component that links Africans with Middle-easterners, it is neither just African nor just Mideastern, but both, however, it is found at a more higher frequency with indigenous SubSaharn Eastern Africans, like the Sandawe and Maasai (13-14%) and Ethiopians (40%) than it is found with West Africans.
For the West African component, it is found in Northern Africans at a median frequency in the following order:
mozabite 16.34%
morocco-s 15.75%
sahara-occ 15.32%
moroccans 14.80%
algeria 13.02%
tunisia 12.88%
morocco-n 11.91%
libya 9.45%
egypt 6.67%
egyptans 6.60%
Ethiopians had it at 10-11%, so likely this component is a kind of pan-African component that has its presence in a variety of African populations due to many millennia of inter-African migrations/interactions.
“this is not seen in other analysis of this type except maybe for some of the South Morroco. “
Many of the analysis out there do not provide you with the direct samples they are using since they include a large number of private samples from participants in 'projects', this analysis is different because I actually provide you with ALL the direct samples and polymorphisms that I employed for this analysis since they are all public, thus you can verify the ADMIXTURE analysis for yourself if you have the requisite software installed.
Etyopsis, first i want to thankyou greatly for this post, its very informative and the Dataset will help me get started on a "New World" Ancestry project, in which it is Vital for the African components to be as broken down as possible, but will also need to keep alot of the populations for people with native ancestry, east asian, south asian, and southeast asian, siberian, south/north euro, mideast and north-african. Do you have any tips/suggestoins for having the Western Bantu and Eastern bantu's cluster form? Or any suggestoins at all will be very welcome, i am Lemba from ABF
ReplyDeleteHi Lemba, from my observations so far, the eastern and Western Bantu clusters don't form on a global level from the current SNPs that are included in my dataset, they do however form from an inter-continental African prespective. See this post for details: Intra African Genome-Wide Analysis
DeleteThe populations you have listed are very widespread globally, so when you include all those populations it becomes more of a global analysis, and thus it may be difficult to split the Eastern from Western Bantu/Niger Kordofani.
So, you could try the following steps,
1) Start with the base intra African dataset, as I have outlined in the post I linked you to above. Include the New World populations in that dataset and see if the Eastern and Western Bantu components are still splitting, there the North Africans can act as a proxy for Eurasian gene-flow to start with.
2) Then add one by one the Eurasian populations you are interested in to that dataset, start with the ones that are furthest away from Africa, Native Americans, East Asians,...... See if the Eastern and Western Bantu clusters are still splitting, if so you can add more populations from Eurasia, but at some point the components will stop splitting I am just not sure at which point it will be.
I have uploaded the base Intra African dataset I use in PLINK format here , you need to create your new world dataset to merge with however, if you haven't done so already. You also need to be mindful of your K selection, just because some components split at a given K, it doesn't necessarily mean they are statistically useful, hence try to utilize the cross-validation error values ADMIXTURE computes for each K run, even though it may take a while for your machine to process.
Hope this helps for you to get started, let me know if you have other questions.
Thanks for the advice!, I just included amerindians into your pan-african dataset to see how it behaves. Will run a K=14 tonight and when i have all the populations in, ill do the K validation you did.
DeleteHow are you merging in 23andme data files? I am using a script Rhazib had on his site, and i was able to generate a .tped and .tfam file. Now my questoin is, after i make that .bed .bim .fam, don't i have to filter only the snp's which these datasets are using?
23andme data suggestions would really help = )
Yes, you have to filter out the SNPs used in the dataset from those in your raw data, to do so, after you have successfully converted your raw data to the PLINK format (.bed,.bim, .fam) run the following commands in PLINK.
DeleteAssuming the dataset is the Africa one I posted earlier, i.e "Africa_Rev4_public"
First, extract the SNPs:
plink --bfile "Africa_Rev4_public" --write-snplist --out "Extracted_SNPs"
This will write a file called "Extracted_SNPs.snplist", in your working folder.
Then, use that new file to extract the SNPs from your raw data, assuming your raw data is named "my_rawdata", use the following:
plink --bfile "my_rawdata" --extract "Extracted_SNPs.snplist" --make-bed --out "my_rawdata_filtered"
This will make new .bed,.bim and .fam files called "my_rawdata_filtered"
Lastly, merge your filtered raw data with the main file you are using ("Africa_Rev4_public" in this case)
plink --bfile "Africa_Rev4_public" --bmerge "my_rawdata_filtered.bed" "my_rawdata_filtered.bim" "my_rawdata_filtered.fam" --make-bed --out "New_file"
This will make new .bed,.bim and .fam files called "New_file", which you can use to run ADMIXTURE with.
One last note, the script that Razib had on his blog did not work well for me to convert 23andME raw data, it had some issues with the no calls, I'm not sure if it worked out for you, in any event I had to write my own code using GNU OCTAVE to convert 23andME raw data to the appropriate .tped and .tfam PLINK formatted files, if you want I can make a separate posting outlining how to do that.......
Wow thanks! this is what i needed. Yes in the SH script i got an error for the tfam, but with the perl script i didn't , although maybe its not being verbose. If you can post your own code i would really appreciate it! Also how do you output the current population list inside a .bed file? For example after i add myself how do i extract the population list .txt out of the .bed
DeleteI have created a post for converting 23andME raw data. Check it out and let me know if it works for you.
DeleteAs far as outputting the current population list you are using, I have a separate but a little more complicated program for correlating fam files with a superset text file, I know exactly what you are saying though, since the .fam files only has a listing of the family and Individual Id's it is hard to say which population the sample belongs to by just looking at it, if you have only added a few samples I suggest you can just manually identify them, and then add them to your original population list manually, the critical thing is that the samples have to be in the same order as the fam file as that is the order ADMIXTURE will report the output of the cluster proportions in the .q file.
Why doesn't the MENA cluster break up?
ReplyDeleteIt breaks up at K=14, even-though the cross validation error at that point is higher than that of K13's, and so the results may not be as reliable as for the K found at the lowest cross validation error. I had attached the K14 results anyway with the other data files in my original post.
DeleteAt K14, the MENA cluster breaks up into a North West African and a South West Asian cluster that peaks in the Tunisians and Bedouins respectively. An additional Polynesian cluster that peaks in the Samoans is also formed while the Pygmy cluster disappears to compensate for it.
The interesting thing however about the MENA cluster breaking up @ K14 into a NW African and SW Asian cluster is that both the components are significantly present in the Ethiopian dataset at about 17% and 30% respectively, the fact that the MENA cluster breaks up into such spatially spread but distinct components, while both components simultaneously appear in the Ethiopian dataset points to its (i.e. MENA component's) relative antiquity in Africa, perhaps a significant portion of it is even older in Africa than it is in the middle-east itself.